Stage 30
Here we can fit the broadband (“white”) light curve (which was created in S20) or spectroscopic light curves (which were created in S21).
Let’s remove the first orbit from every visit and the first exposure from every orbit as they are typically strongly affected by instrument systematics:
We can choose if we also want to run an MCMC using the emcee package after running the least squares routine:
For the MCMC, let’s do a quick, small number of samples with an number of walkers at least greater than twice the numbers of free parameters.
Note: The user can also use dynesty, a nested sampler, instead of emcee.
Let’s use the following model:
‘constant’: a normalization constant
‘upstream_downstream’: accounts for the forward and reverse scanning effect which creates an offset in the measured flux
‘model_ramp’: a ramp for every orbit
‘polynomial1’: a slope over the visit
‘transit’: a BATMAN transit model
Additional functions are listed on the models page.
Let’s have the following free parameters:
t0_0, rp_0, u1_0, c_0, c_1, v_0, v_1, r1_0, r1_1, r2_0, r2_1, scale_0, scale_1
Other important parameters (per, ars, inc) are fixed to the literature values.
The user can set in the pcf whether the uncertainties should be rescaled to achieve a reduced chi2 of unity. An alternative method is using an additional free parameter which rescales the uncertainties at every step of the sampler. This model is called uncmulti.
White light curve fit
Here’s the fit_par.txt file which was used in this example to fit the white light curve:
#parameter fixed tied value lo_lim lo_val hi_lim hi_val prior p1 p2 step_size
per True -1 1.5804046 True 1.55 True 1.6 X 2.25314 2e-05 1e-07
t0 False -1 0.18 True 0.16 True 0.2 U 0.0 0.3 0.001
t_secondary True -1 0.0 False 0.0 False 0.0 X 0.0 0.0 0.0
w True -1 90.0 False 0.0 False 0.0 X 0.0 0.0 0.0
a True -1 15.23 True 14.0 True 16.0 X 4.98 0.05 0.02
inc True -1 89.1 True 88.0 True 89.9 X 85.3 0.2 0.02
rp False -1 0.116 True 0.05 True 0.2 U 0.01 0.3 0.001
fp True -1 0.0 False 0.0 False 0.002 X 0.0 1.0 0.0
u1 False -1 0.29 True 0.0 True 0.6 U 0.0 1.0 0.01
u2 True -1 0.0 False 0.0 False 0.0 X 0.0 0.0 0.0
ecc True -1 0.0 False 0.0 False 0.0 X 0.0 0.0 0.0
c False 0 8.37 True 6.0 True 8.9 X 6.7 7.0 0.001
c False 1 8.37 True 6.0 True 8.9 X 6.7 7.0 0.001
v False 0 -1e-06 False -0.0001 False -1e-08 X -7e-06 -1e-06 1e-05
v False 1 -1e-06 False -0.0001 False -1e-08 X -7e-06 -1e-06 1e-05
v2 True -1 0.0 True 0.0 True 1.0 X 0.0 0.0 1e-11
r1 False 0 0.1 False 0.0 False 1.0 U -10.0 10.0 0.01
r1 False 1 0.1 False 0.0 False 1.0 U -10.0 10.0 0.01
r2 False 0 0.0 False 5.5 False 7.5 U -100.0 100.0 0.1
r2 False 1 0.0 False 5.5 False 7.5 U -100.0 100.0 0.1
r3 True 0 0.0 False -1.0 False 1.0 U -10.0 10.0 0.001
r3 True 1 0.0 False -1.0 False 1.0 U -10.0 10.0 0.001
scale False 0 0.0 False -0.01 False 0.01 U -0.1 0.1 0.002
scale False 1 0.0 False -0.01 False 0.01 U -0.1 0.1 0.002
uncmulti_val True -1 1.0 False 1.0 False 7.0 U 0.1 10.0 0.01
If a parameter is free and not jointly shared across visits, the user has to repeat the parameter in the fit_par.txt file as shown above. Furthermore, the visit number must be added in the “tied” column. tied = -1 means that the parameters is shared across all visits, which makes sense for orbital parameters but less for systematics like the normalization constant.
Let’s run the white light curve fit now:
Successfully reloaded meta file
Starting s30
White light curve fit will be performed
using most recent s20 run: 2022-10-20_13-36-21
Identified file(s) for fitting: ['./run_2022-10-20_13-09-52_GJ1214_Hubble13021//extracted_lc/2022-10-20_13-36-21/lc_white.txt']
****** File: 1/1
Removed 8 exposures because they were the first exposures in the orbit.
Removed 34 exposures because they were the first orbit in the visit.
median log10 raw flux: 8.429980180844982
The highest amount of exposures in an orbit is 18
Number of free parameters: 13
Names of free parameters: ['t0', 'rp', 'u1', 'c', 'c', 'v', 'v', 'r1', 'r1', 'r2', 'r2', 'scale', 'scale']
The predicted rms is 63.68
*STARTS LEAST SQUARED*
Runs MPFIT...
/home/zieba/Desktop/Projects/Open_source/PACMAN/src/pacman/lib/model.py:82: RuntimeWarning: divide by zero encountered in true_divide
self.data_nosys = data.flux/self.model_sys
/home/zieba/Desktop/Projects/Open_source/PACMAN/src/pacman/lib/model.py:83: RuntimeWarning: divide by zero encountered in true_divide
self.norm_flux = data.flux/self.model
t0_0 1.8276e-01 1.0250e-05
rp_0 1.1615e-01 8.2543e-05
u1_0 2.6010e-01 5.0892e-03
c_0 8.4302e+00 1.6253e-05
c_1 8.4300e+00 1.6509e-05
v_0 -1.2284e-06 1.1059e-07
v_1 -2.5289e-07 1.1064e-07
r1_0 6.7861e-02 6.1244e-03
r1_1 6.5916e-02 5.4471e-03
r2_0 6.7533e+00 3.2065e-02
r2_1 6.6605e+00 2.9597e-02
scale_0 4.1908e-03 1.7515e-05
scale_1 4.1977e-03 1.7513e-05
rms, chi2red = 122.480430626872 4.206812618533217
Saved white_systematics.txt file
['t0', 'rp', 'u1', 'c0', 'c1', 'v0', 'v1', 'r10', 'r11', 'r20', 'r21', 'scale0', 'scale1']
*STARTS MCMC*
Runs MPFIT...
/home/zieba/Desktop/Projects/Open_source/PACMAN/src/pacman/lib/model.py:82: RuntimeWarning: divide by zero encountered in true_divide
self.data_nosys = data.flux/self.model_sys
/home/zieba/Desktop/Projects/Open_source/PACMAN/src/pacman/lib/model.py:83: RuntimeWarning: divide by zero encountered in true_divide
self.norm_flux = data.flux/self.model
t0_0 1.8276e-01 2.1023e-05
rp_0 1.1615e-01 1.6930e-04
u1_0 2.6010e-01 1.0438e-02
c_0 8.4302e+00 3.3335e-05
c_1 8.4300e+00 3.3861e-05
v_0 -1.2284e-06 2.2682e-07
v_1 -2.5289e-07 2.2692e-07
r1_0 6.7861e-02 1.2561e-02
r1_1 6.5916e-02 1.1172e-02
r2_0 6.7533e+00 6.5767e-02
r2_1 6.6605e+00 6.0704e-02
scale_0 4.1908e-03 3.5925e-05
scale_1 4.1977e-03 3.5919e-05
rms, chi2red = 122.48043062689561 1.0000000000003877
Run emcee...
100%|#######################| 4000/4000 [00:44<00:00, 90.09it/s]
Saved white_systematics.txt file for mcmc run
Saved fit_results.txt file
Finished s30
There are several plots created then:
The raw light curve:
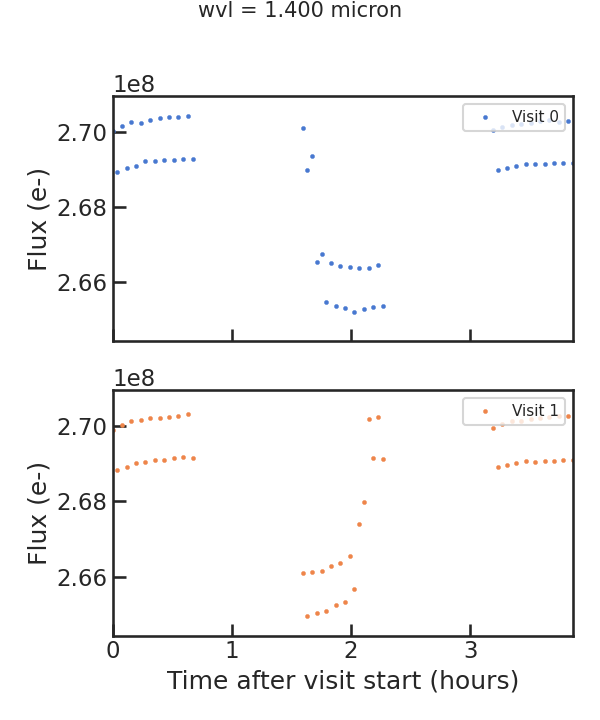
** From the least squares routine **
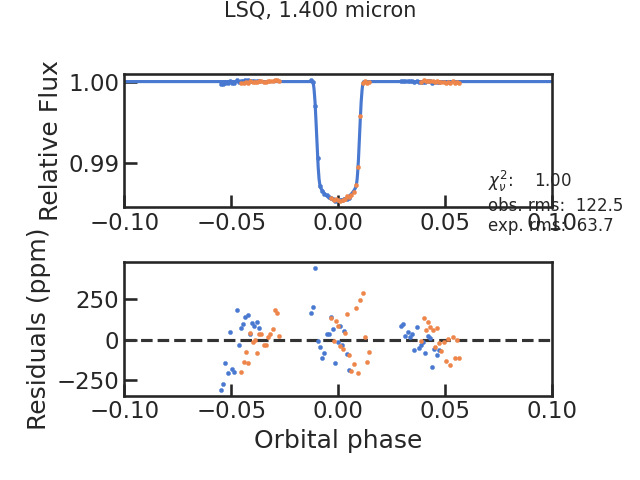
The fitted light curve without the systematics:
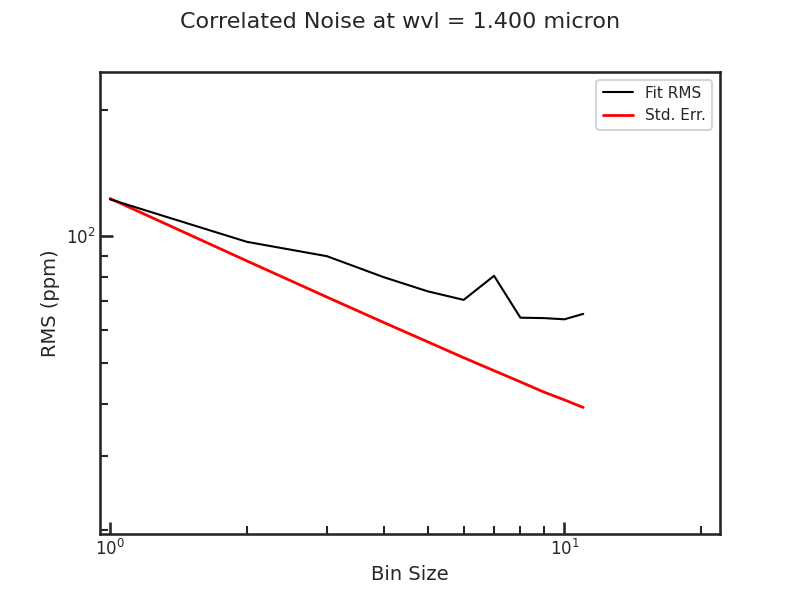
The Allan deviation plot:
** Using emcee **
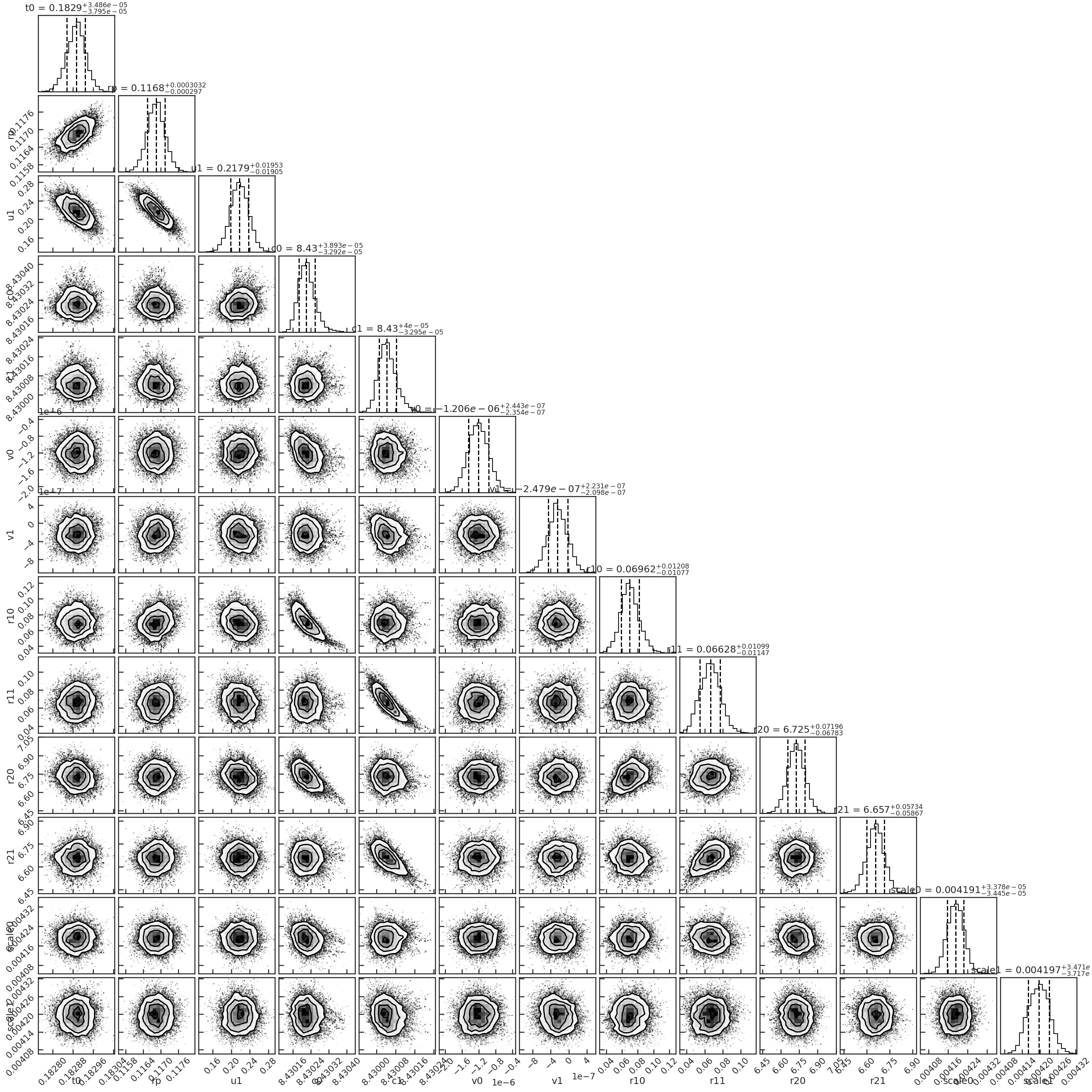
The fitted light curve without the systematics:
MCMC chains with burn-in:
MCMC chains without burn-in
Corner plot from the MCMC:
Spectroscopic light curve fit
Here’s the fit_par.txt file which was used in this example to fit the spectroscopic light curves:
#parameter fixed tied value lo_lim lo_val hi_lim hi_val prior p1 p2 step_size
per True -1 1.5804046 True 1.55 True 1.6 X 2.25314 2e-05 1e-07
t0 False -1 0.18 True 0.16 True 0.2 U 0.0 0.3 0.001
t_secondary True -1 0.0 False 0.0 False 0.0 X 0.0 0.0 0.0
w True -1 90.0 False 0.0 False 0.0 X 0.0 0.0 0.0
a True -1 15.23 True 14.0 True 16.0 X 4.98 0.05 0.02
inc True -1 89.1 True 88.0 True 89.9 X 85.3 0.2 0.02
rp False -1 0.116 True 0.05 True 0.2 U 0.01 0.3 0.001
fp True -1 0.0 False 0.0 False 0.002 X 0.0 1.0 0.0
u1 False -1 0.29 True 0.0 True 0.6 U 0.0 1.0 0.01
u2 True -1 0.0 False 0.0 False 0.0 X 0.0 0.0 0.0
ecc True -1 0.0 False 0.0 False 0.0 X 0.0 0.0 0.0
c False 0 8.37 True 6.0 True 8.9 X 6.7 7.0 0.001
c False 1 8.37 True 6.0 True 8.9 X 6.7 7.0 0.001
v False 0 -1e-06 False -0.0001 False -1e-08 X -7e-06 -1e-06 1e-05
v False 1 -1e-06 False -0.0001 False -1e-08 X -7e-06 -1e-06 1e-05
v2 True -1 0.0 True 0.0 True 1.0 X 0.0 0.0 1e-11
r1 False 0 0.1 False 0.0 False 1.0 U -10.0 10.0 0.01
r1 False 1 0.1 False 0.0 False 1.0 U -10.0 10.0 0.01
r2 False 0 0.0 False 5.5 False 7.5 U -100.0 100.0 0.1
r2 False 1 0.0 False 5.5 False 7.5 U -100.0 100.0 0.1
r3 True 0 0.0 False -1.0 False 1.0 U -10.0 10.0 0.001
r3 True 1 0.0 False -1.0 False 1.0 U -10.0 10.0 0.001
scale False 0 0.0 False -0.01 False 0.01 U -0.1 0.1 0.002
scale False 1 0.0 False -0.01 False 0.01 U -0.1 0.1 0.002
uncmulti_val True -1 1.0 False 1.0 False 7.0 U 0.1 10.0 0.01
Successfully reloaded meta file
Starting s30
Spectroscopic light curve fit(s) will be performed
using most recent s21 run: ./run_2022-10-20_13-09-52_GJ1214_Hubble13021//extracted_sp/bins11_2022-10-20_13-39-31
Identified file(s) for fitting: ['./run_2022-10-20_13-09-52_GJ1214_Hubble13021//extracted_sp/bins11_2022-10-20_13-39-31/speclc1.158.txt', './run_2022-10-20_13-09-52_GJ1214_Hubble13021//extracted_sp/bins11_2022-10-20_13-39-31/speclc1.204.txt', './run_2022-10-20_13-09-52_GJ1214_Hubble13021//extracted_sp/bins11_2022-10-20_13-39-31/speclc1.250.txt', './run_2022-10-20_13-09-52_GJ1214_Hubble13021//extracted_sp/bins11_2022-10-20_13-39-31/speclc1.296.txt', './run_2022-10-20_13-09-52_GJ1214_Hubble13021//extracted_sp/bins11_2022-10-20_13-39-31/speclc1.342.txt', './run_2022-10-20_13-09-52_GJ1214_Hubble13021//extracted_sp/bins11_2022-10-20_13-39-31/speclc1.389.txt', './run_2022-10-20_13-09-52_GJ1214_Hubble13021//extracted_sp/bins11_2022-10-20_13-39-31/speclc1.435.txt', './run_2022-10-20_13-09-52_GJ1214_Hubble13021//extracted_sp/bins11_2022-10-20_13-39-31/speclc1.481.txt', './run_2022-10-20_13-09-52_GJ1214_Hubble13021//extracted_sp/bins11_2022-10-20_13-39-31/speclc1.527.txt', './run_2022-10-20_13-09-52_GJ1214_Hubble13021//extracted_sp/bins11_2022-10-20_13-39-31/speclc1.573.txt', './run_2022-10-20_13-09-52_GJ1214_Hubble13021//extracted_sp/bins11_2022-10-20_13-39-31/speclc1.619.txt']
****** File: 1/11
Removed 8 exposures because they were the first exposures in the orbit.
Removed 34 exposures because they were the first orbit in the visit.
median log10 raw flux: 6.246127147951612
The highest amount of exposures in an orbit is 18
Number of free parameters: 13
Names of free parameters: ['t0', 'rp', 'u1', 'c', 'c', 'v', 'v', 'r1', 'r1', 'r2', 'r2', 'scale', 'scale']
The predicted rms is 247.28
*STARTS LEAST SQUARED*
Runs MPFIT...
/home/zieba/Desktop/Projects/Open_source/PACMAN/src/pacman/lib/model.py:82: RuntimeWarning: divide by zero encountered in true_divide
self.data_nosys = data.flux/self.model_sys
/home/zieba/Desktop/Projects/Open_source/PACMAN/src/pacman/lib/model.py:83: RuntimeWarning: divide by zero encountered in true_divide
self.norm_flux = data.flux/self.model
t0_0 1.8281e-01 3.9907e-05
rp_0 1.1644e-01 3.2115e-04
u1_0 2.6678e-01 1.9851e-02
c_0 6.2463e+00 4.7835e-05
c_1 6.2461e+00 4.8850e-05
v_0 -8.9100e-07 4.2938e-07
v_1 -4.6203e-07 4.2955e-07
r1_0 9.3385e-02 3.1389e-02
r1_1 8.8542e-02 3.2951e-02
r2_0 6.8805e+00 1.2962e-01
r2_1 6.9881e+00 1.4549e-01
scale_0 4.1980e-03 6.8081e-05
scale_1 4.0987e-03 6.8061e-05
rms, chi2red = 287.29232332160944 1.5360471033190013
['t0', 'rp', 'u1', 'c0', 'c1', 'v0', 'v1', 'r10', 'r11', 'r20', 'r21', 'scale0', 'scale1']
*STARTS MCMC*
Runs MPFIT...
/home/zieba/Desktop/Projects/Open_source/PACMAN/src/pacman/lib/model.py:82: RuntimeWarning: divide by zero encountered in true_divide
self.data_nosys = data.flux/self.model_sys
/home/zieba/Desktop/Projects/Open_source/PACMAN/src/pacman/lib/model.py:83: RuntimeWarning: divide by zero encountered in true_divide
self.norm_flux = data.flux/self.model
t0_0 1.8281e-01 4.9460e-05
rp_0 1.1644e-01 3.9802e-04
u1_0 2.6678e-01 2.4603e-02
c_0 6.2463e+00 5.9285e-05
c_1 6.2461e+00 6.0543e-05
v_0 -8.9100e-07 5.3217e-07
v_1 -4.6203e-07 5.3237e-07
r1_0 9.3385e-02 3.8903e-02
r1_1 8.8542e-02 4.0838e-02
r2_0 6.8805e+00 1.6065e-01
r2_1 6.9881e+00 1.8032e-01
scale_0 4.1980e-03 8.4377e-05
scale_1 4.0987e-03 8.4353e-05
rms, chi2red = 287.29232332158693 0.9999999999998428
Run emcee...
100%|███████████████████████████████████████████████████████████████████████████████████████████████████| 4000/4000 [00:43<00:00, 92.36it/s]
****** File: 2/11
Finished s30
Most plots which are created during the white light curve fit will be also created after running the spectroscopic fits. Let’s look at some examples:
The first raw spectroscopic light curve:
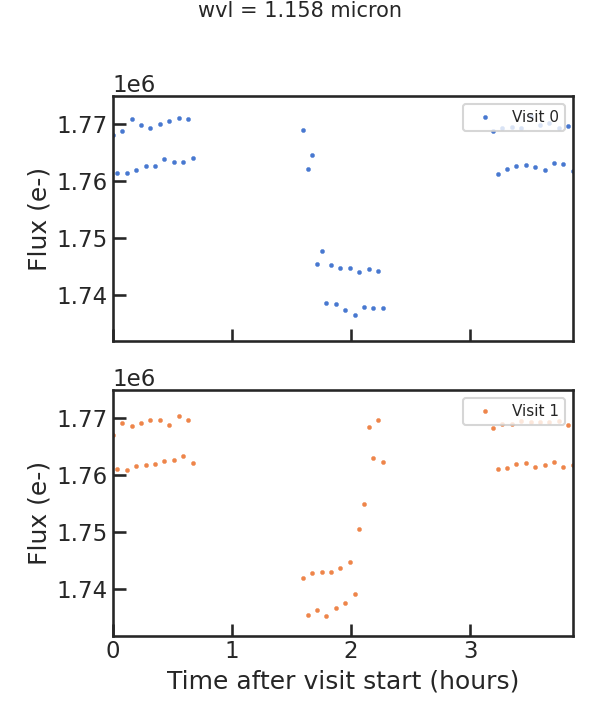
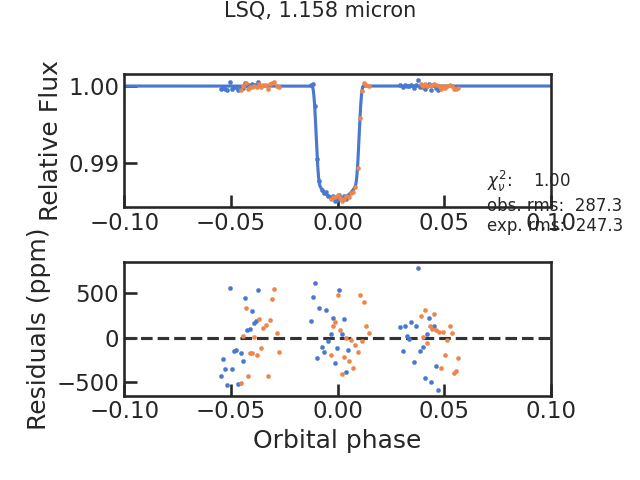
** Using least squared **
The fitted spectroscopic light curve without the systematics:
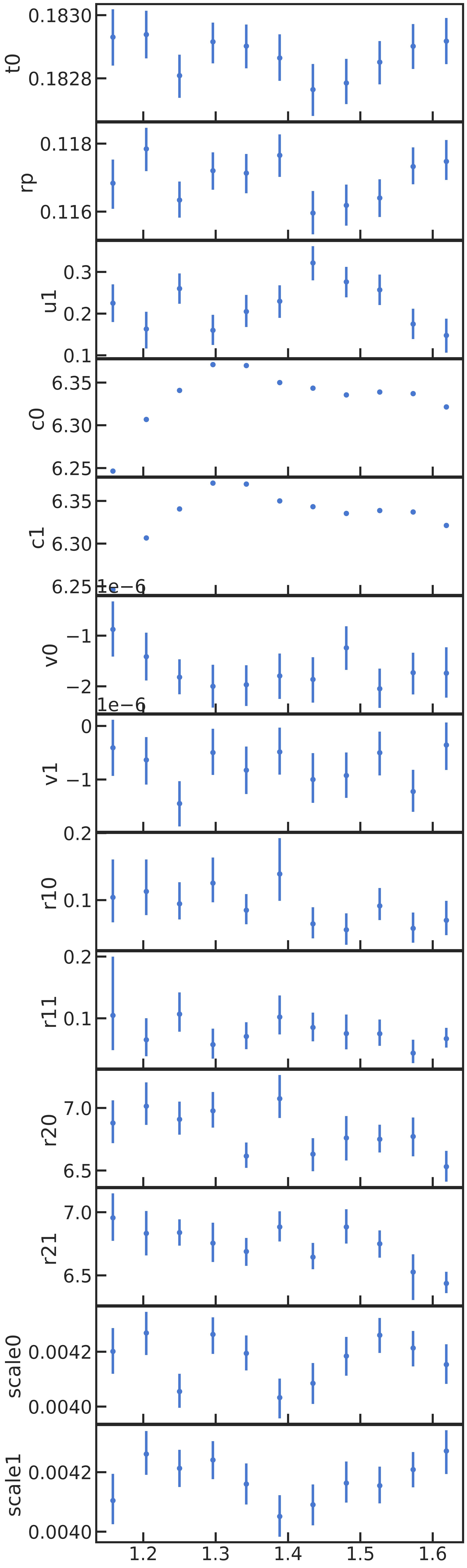
All fitted parameters as a function of wavelength:
** Using emcee **
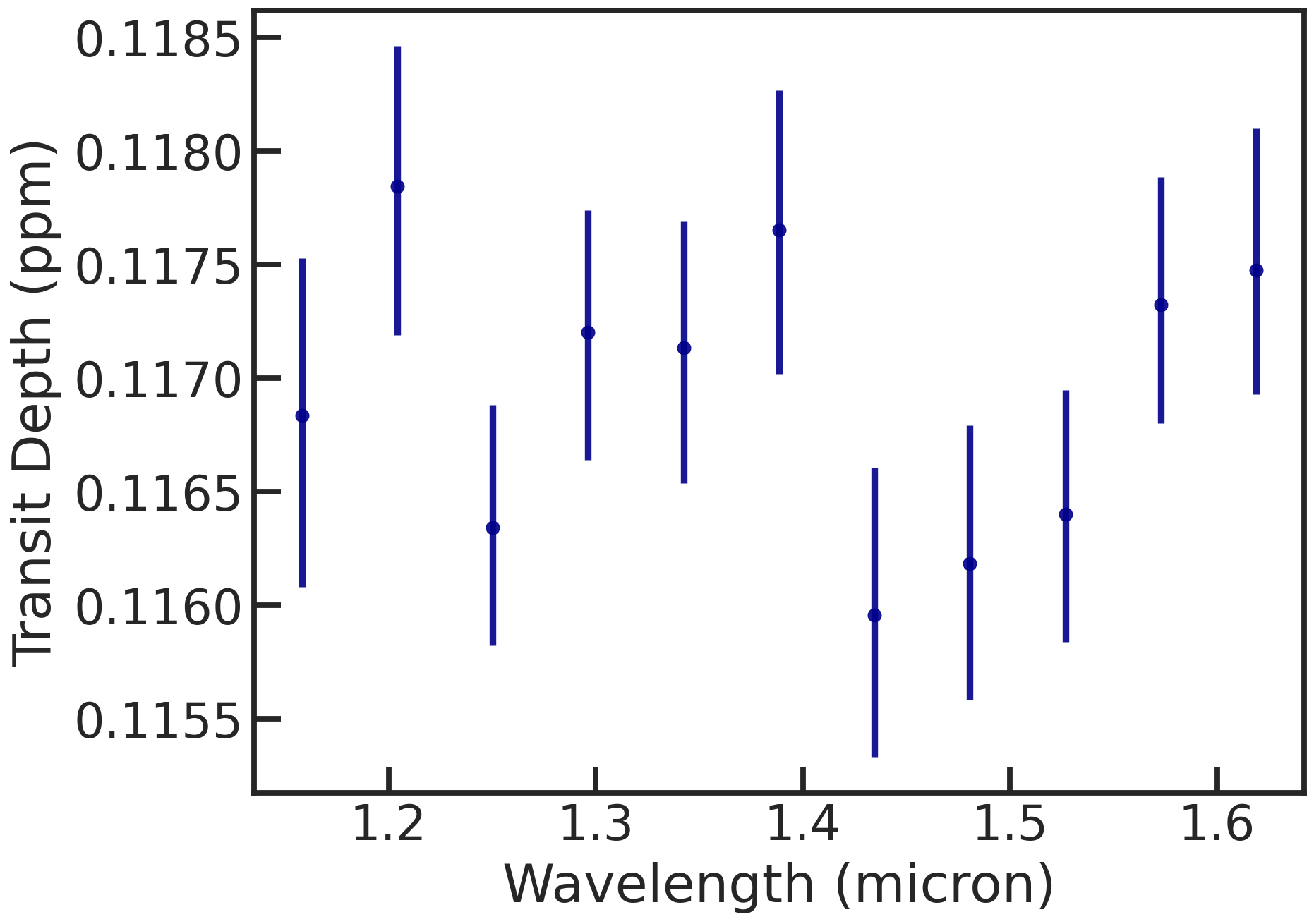
The spectrum (rprs vs wavelength):