Download data (alternatives)
There are different ways to download the data. Let’s have a look at them by downloading the data taken in HST Program GO 13021.
Note
PACMAN can currently just work with files with an
imaextension, so you want to select these.imais an intermediate data product standing for calibrated intermediate IR multiaccum image. From the WFC3 data handbook (Types of WFC3 Files): “For the IR detector, an intermediate MultiAccum (ima) file is the result after all calibrations are applied (dark subtraction, linearity correction, flat fielding, etc.) to all of the individual readouts of the IR exposure.”
Using the program’s site
Go to the new HST MAST search tool.
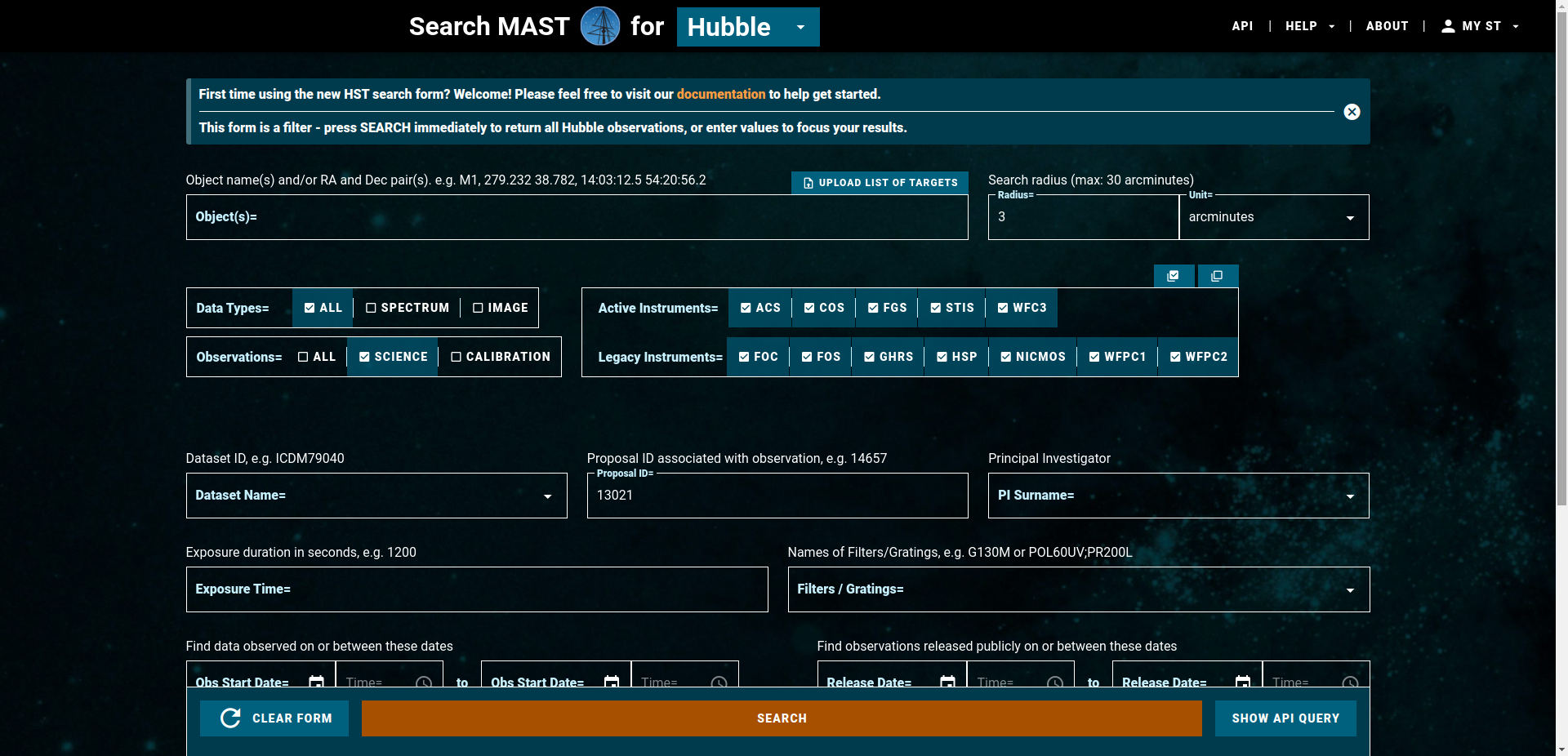
Enter 13021 for Proposal ID and click on search.
All files associated with the program will be then shown in the following table.
One of the columns is called Proposal ID.
You can see that all of them say 13021 because that’s what we wanted. Click on any of the numbers saying 13021.
You should get to program site.
It lists interesting information to the program like papers associated with the program and also (further down on the website) all files taken in this program.
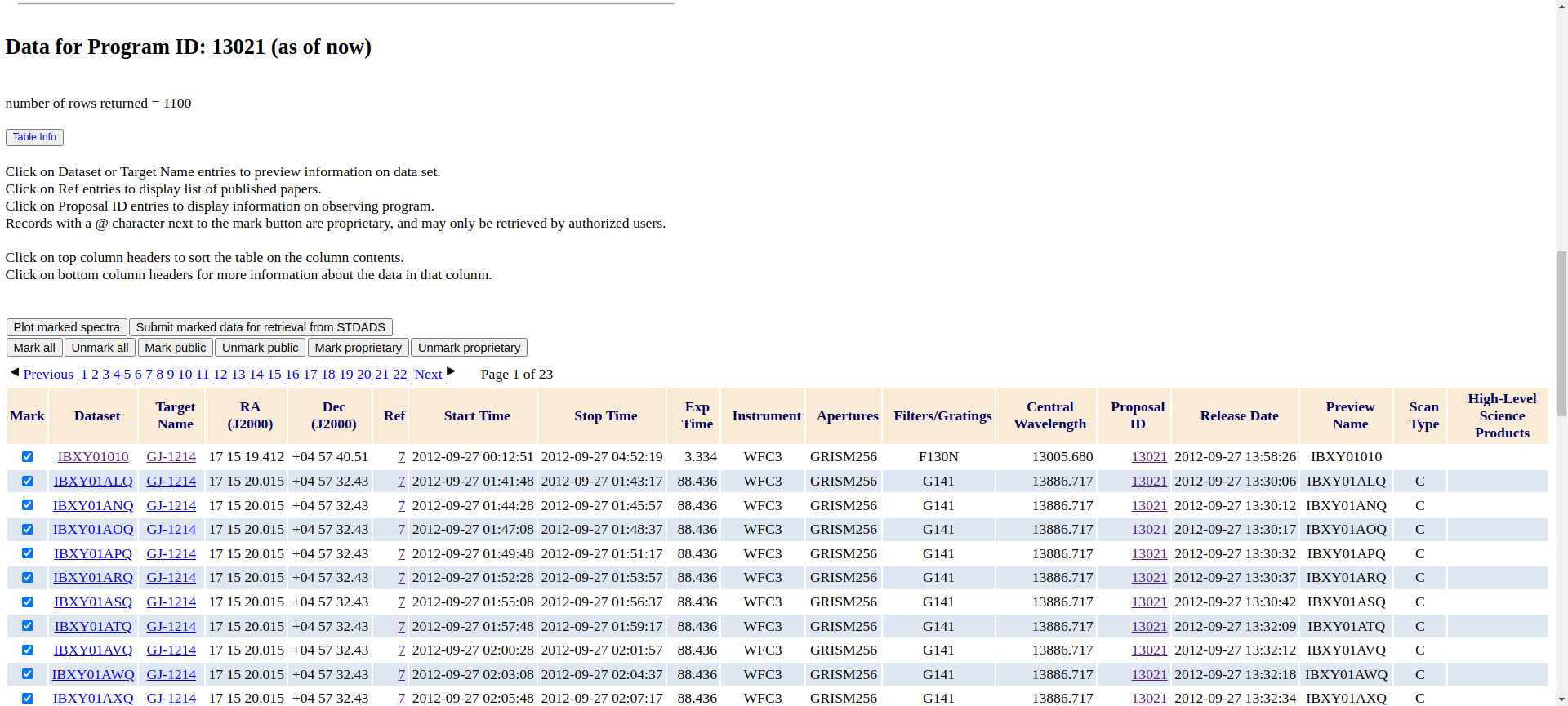
Let’s click on Mark all right above the table. And then on Submit marked data for retrieval from STDADS.
A new page will load where you can finally download your data.
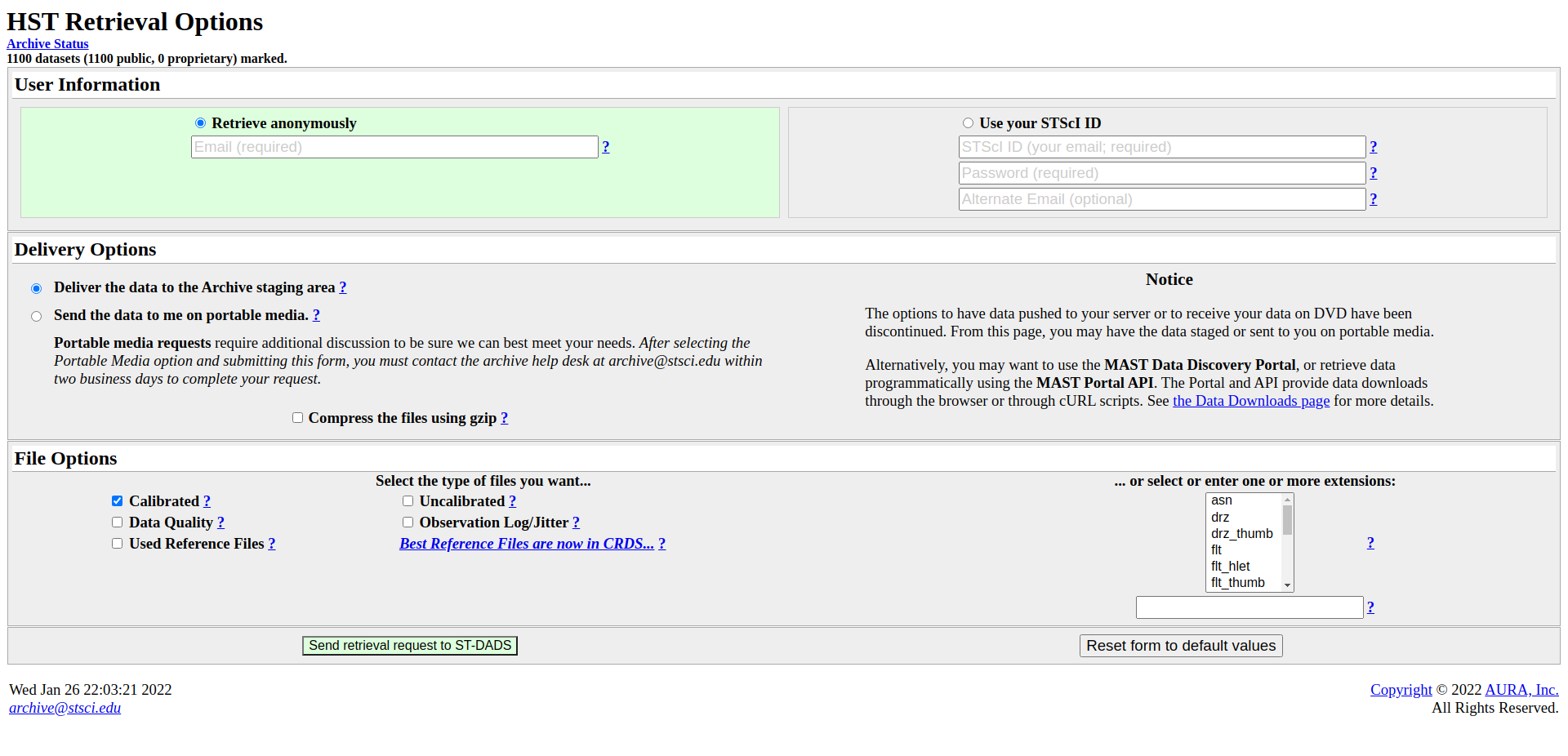
Enter your email address or use your STScI log in. (If the data you want to download is still proprietary, you will have to use the log-in)
File type: Calibrated data (should be marked already)
Extension (bottom right): ima
Click Send retrieval request to ST-DADS
You should then get more instructions per email.
You can find more information how to download the files with ftp here: Retrieval Options, Retrieving IUE Data via FTP and MAST FTP Service.
You can access the data using the ftplib module in python. Here’s an example script which you can also find on GitHub. It will save download the data into the current directory.
import ftplib
import getpass
ftps = ftplib.FTP_TLS('archive.stsci.edu')
#https://archive.stsci.edu/ftp.html
#If the data you are downloading is not exclusive access (that is, is publicly accessible),
#then you can use an anonymous connection, with "anonymous" as the username and your email address as the password.
#(We don't do anything with the email address, so you can enter something else if you like.)
user = getpass.getpass(prompt='Enter email address (if anonymous -> enter "anonymous"): ', stream=None)
passwd = getpass.getpass(prompt='Enter password (if anonymous -> enter email address): ', stream=None)
ftps.login(user=user, passwd=passwd)
ftps.prot_p() # This is a really good idea :)
ftps.cwd('stage')
print(ftps.dir())
ftps.cwd('anonymous/anonymous12345') # can be found in the email
filenames = ftps.nlst()
print(filenames)
for filename in filenames:
print("getting " + filename)
with open(filename, 'wb') as fp:
ftps.retrbinary('RETR {}'.format(filename), fp.write)
If necessary, unpack everything and move these fits files into a new directory. This data directory should then contain all the downloaded .fits files. You will have to specify the location of this “data directory” then in the pcf file.
If the user has all files of the 15 visits in the data directory, they can use for example which_visits = [13,14] in the pcf to only analyze the last two visits.
Using the new MAST search tool
An alternative is to download it directly after searching for the program files with the new HST MAST search tool. Note that the website only allows you to mark a maximum of 100 observations to download. You can also download the files by using the old HST MAST search tool which does not have this limitation but might be retired soon.
Because the program has more than 1000 files, this would take some time. So let’s only download the last two visits which were taken in August of 2013 (2013-08-12 and 2013-08-20). Set OBS START DATE = 2013-08-12 and as before, enter 13021 for Proposal ID and click on search.
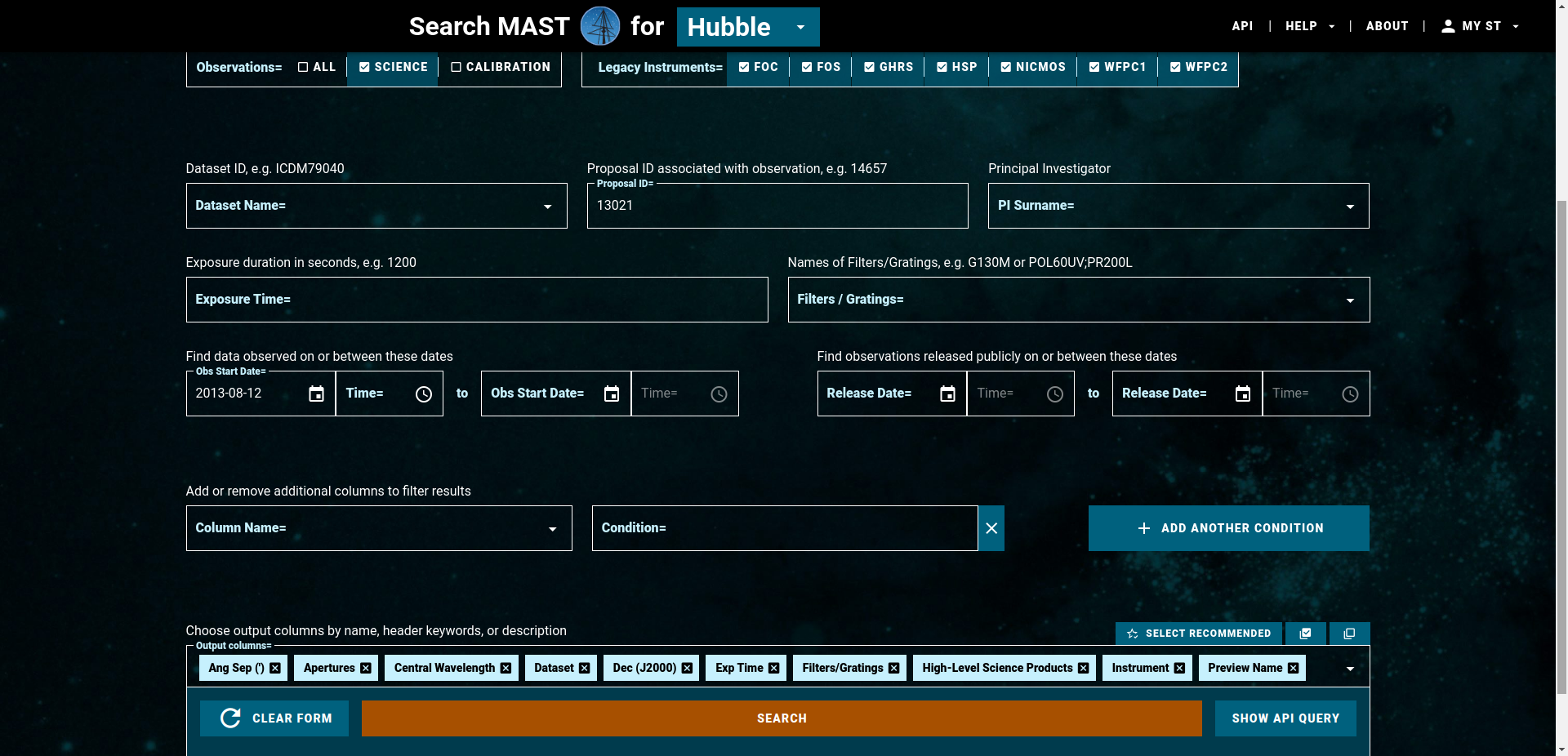
All files associated with the program taken during the last two visits will be then shown in the following table. Go to the buttom right and seclect Rows per Page = 100, then select the 100 files and click on DOWNLOAD DATA (100 DATASETS). A small window opens on the website. Go to the EXTENSIONS drop down list and only select ima. Click on Start DOWNLAÒAD
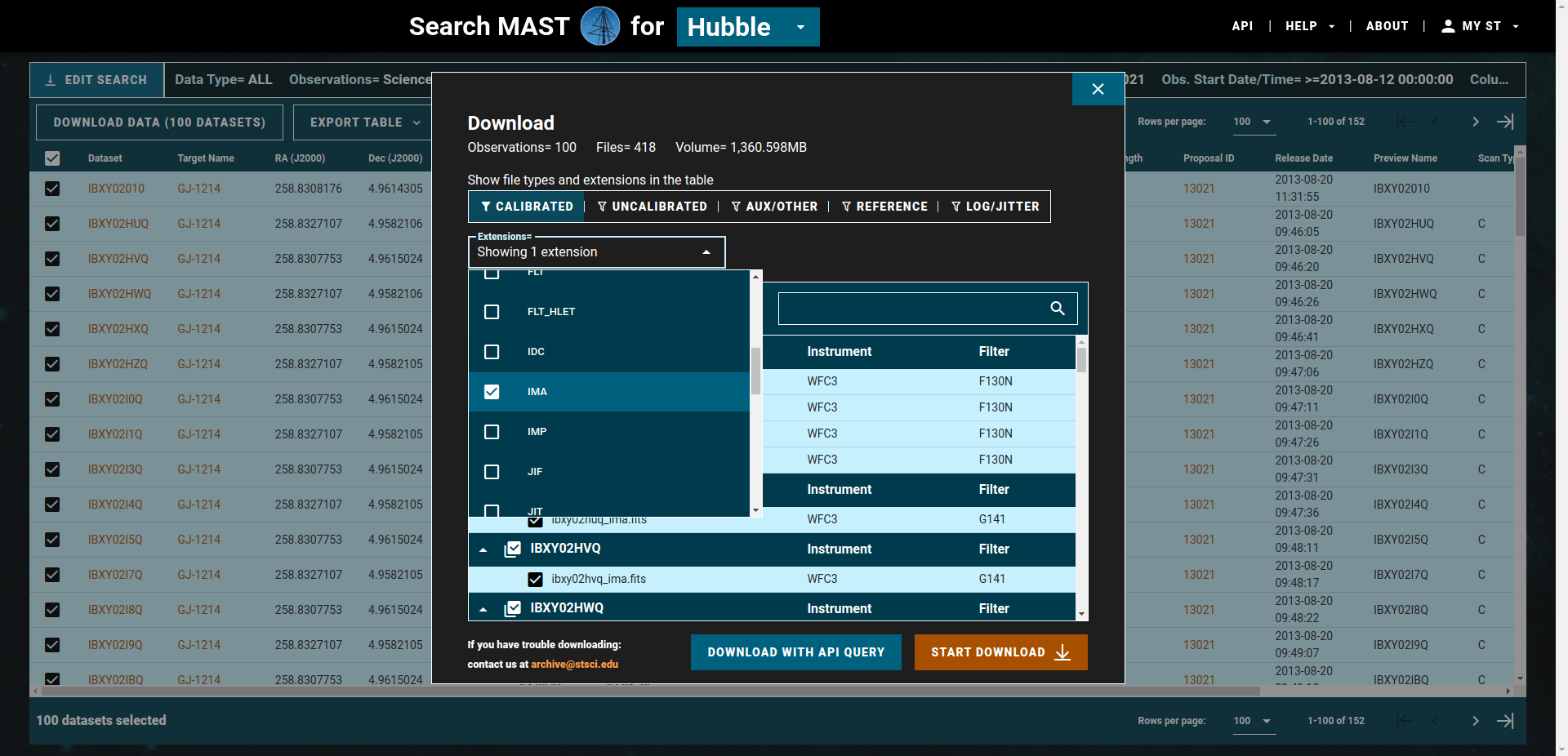
It will download the ima files associated with these 100 datasets as a zip file. After that, do the same for the remaining 52 DATASETS.
Finally, unpack everything and move these fits files into a new directory. This data directory should then just contain the downloaded .fits files. You will have to specify the location of this “data directory” then in the pcf file.